Photographic Explorations

Black aspergilli represent an important group of filamentous fungi. This include species such as Aspergillus niger, which is an important industrial workhorse used for enzyme production as well as being a prolific mycotoxin producer when growing on grapes. In this review, researchers at DTU Bioengineering and CeMiSt give an update on all secondary metabolites from this group of fungi that have been linked to their corresponding genomic biosynthetic gene clusters.
Reference
Front cover of Natural Product Reports Issue 2, 2023
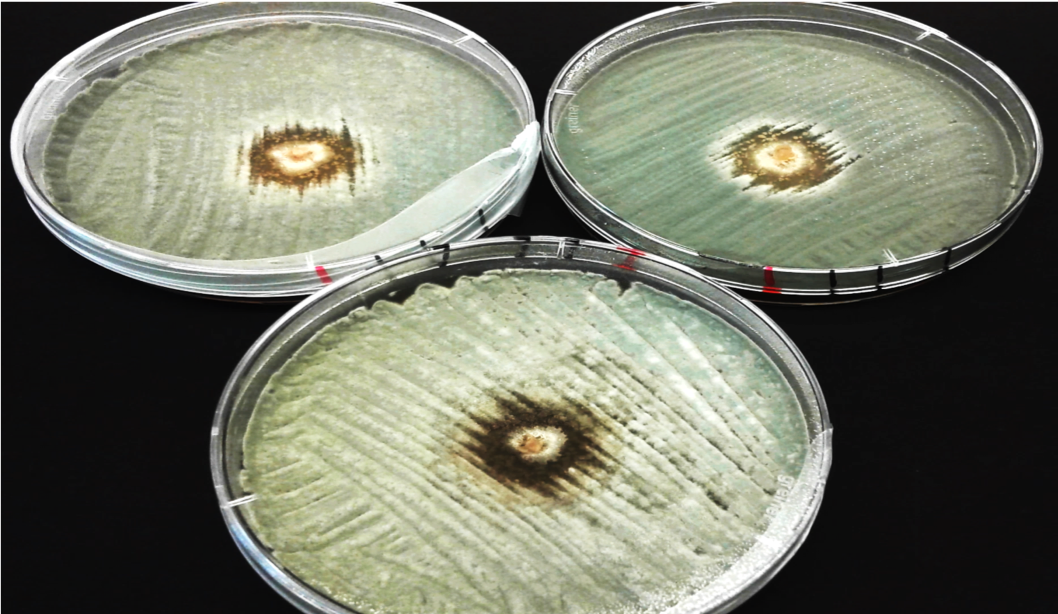
CeMiSt researchers looking for answers on the function of microbial secondary metabolites in natural microbial communities, found a bacterium, Bacillus velezensis that produces a combination of two small chemicals called lipopeptides, which inhibit the growth of different harmful fungi. In particular, the strain B. velezensis can inhibit the human pathogen Candida albicans and its biofilms. This isolate also inhibits the apple-infecting fungus Penicillium expansum. Genome analysis of different Bacillus species revealed that these features of B. velezensis are partly explained by the presence of secondary metabolite gene clusters encoding for the synthesis apparatus of the compounds. The CeMiSt research team demonstrated that the capacity of the bacterium to fight fungal pathogens is related to the presence of lipopeptides. The work included chemical detection methods, genetic analysis of the bacterial strain and traditional microbiological techniques and biofilm assays to reveal these features. The study highlights how CeMiSt stimulates interdisciplinary collaborations resulting in exploitable findings.
Reference
Depiction of secondary metabolites and antifungal activity of Bacillus velezensis DTU001
Azodyrecins- compounds with a rare chemical structure were discovered by CeMiSt scientists. The compounds are produced by a soil bacterium, Streptomyces, isolated from the Danish UNESCO world heritage site Jægersborg Deerpark (DA: Dyrehaven).
Refrences
Azodyrecins A–C: Azoxides from a Soil-Derived Streptomyces Species
Chemistry and Biology of Natural Azoxy Compounds
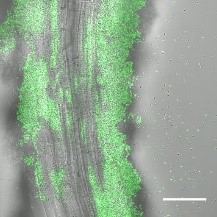
Do microbial secondary metabolites affect the ability of microbes to colonize and establish in new environments?
This basic scientific question has by CeMiSt in Professor Á Kovács’ team that with collaborators have published their findings in the Biofilm journal. In nature, bacterial communities often grow attached to a surface building up thin layers, called biofilms, made of millions of bacterial cells. Professor Kovács and collaborators hypothesized that secondary metabolites released by Bacilli, a group of soil bacteria, had an essential role helping them colonize and grow on surfaces such as plant roots. Surprisingly the team found quite the opposite. When these bacteria, originally isolated from the soil of Dyrehaven, are devoid of the secondary metabolite genes thought to be essential for colonizing surfaces, they saw no difference compared to the wild type. The phenomenon can be observed under the microscope in bacteria producing a fluorescent protein that allows to track them growing along the plant roots. The study exemplifies how little knowledge we have about the role of microbial secondary metabolites in nature. CeMiSt is committed to explore the roles of microbial secondary metabolites in natural microbial systems.
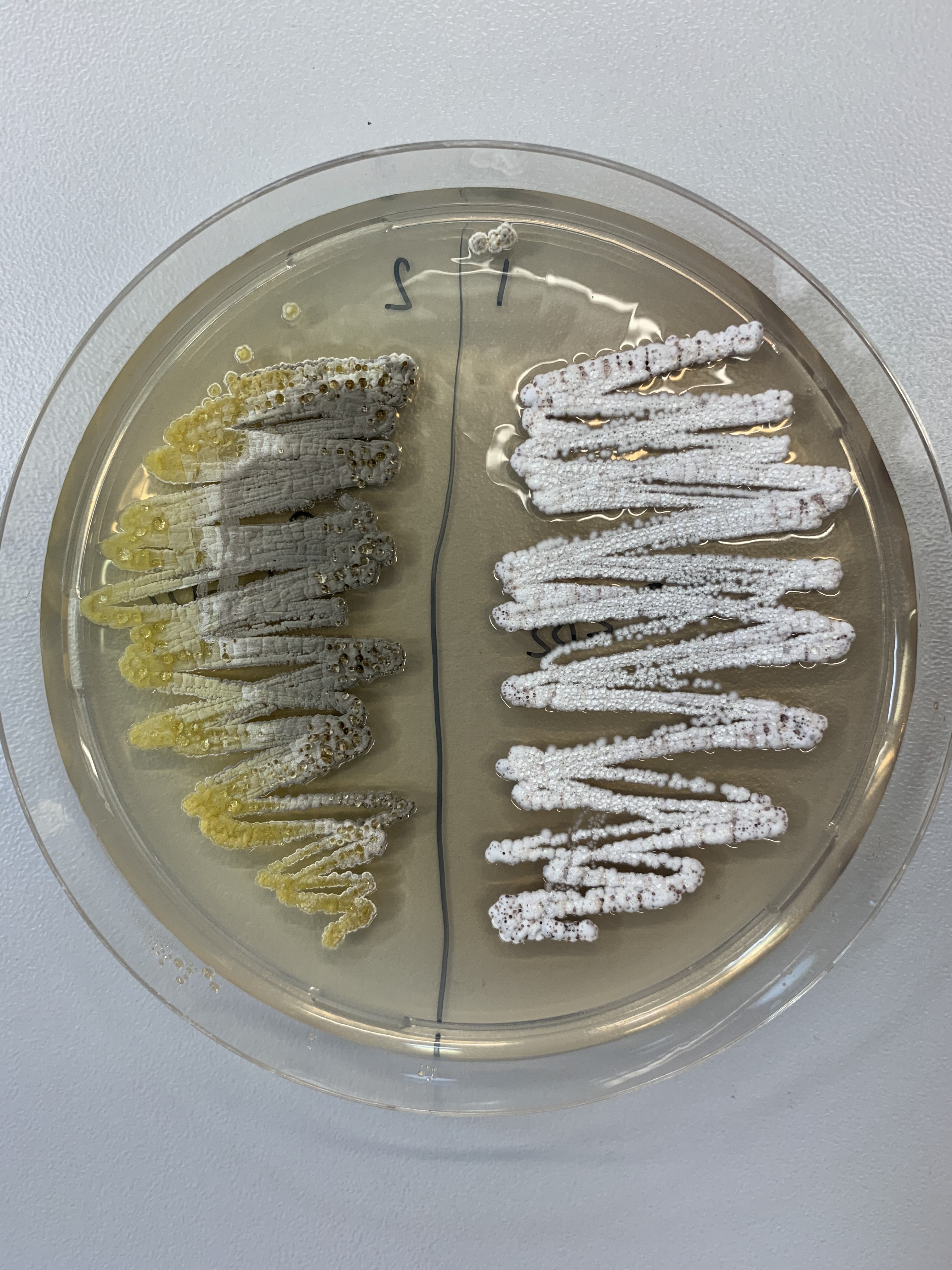
A CeMiSt team has sequenced the complete genomes of 13 Bacillus species from Germany and Denmark whose microbial ecosystems are being investigated. The researchers have looked at the potential secondary metabolite gene cluster of all species using specialized bioinformatic tools. All strains carried genes encoding for the biosynthesis of secondary metabolites surfactin, bacillaene, bacillibactin and pipastatin. This study is a step forward to understand what could possibly be the role of these metabolites in the natural niches of Bacilli. It also highlights the microbial and genetic diversity of one of the center’s model ecosystem.
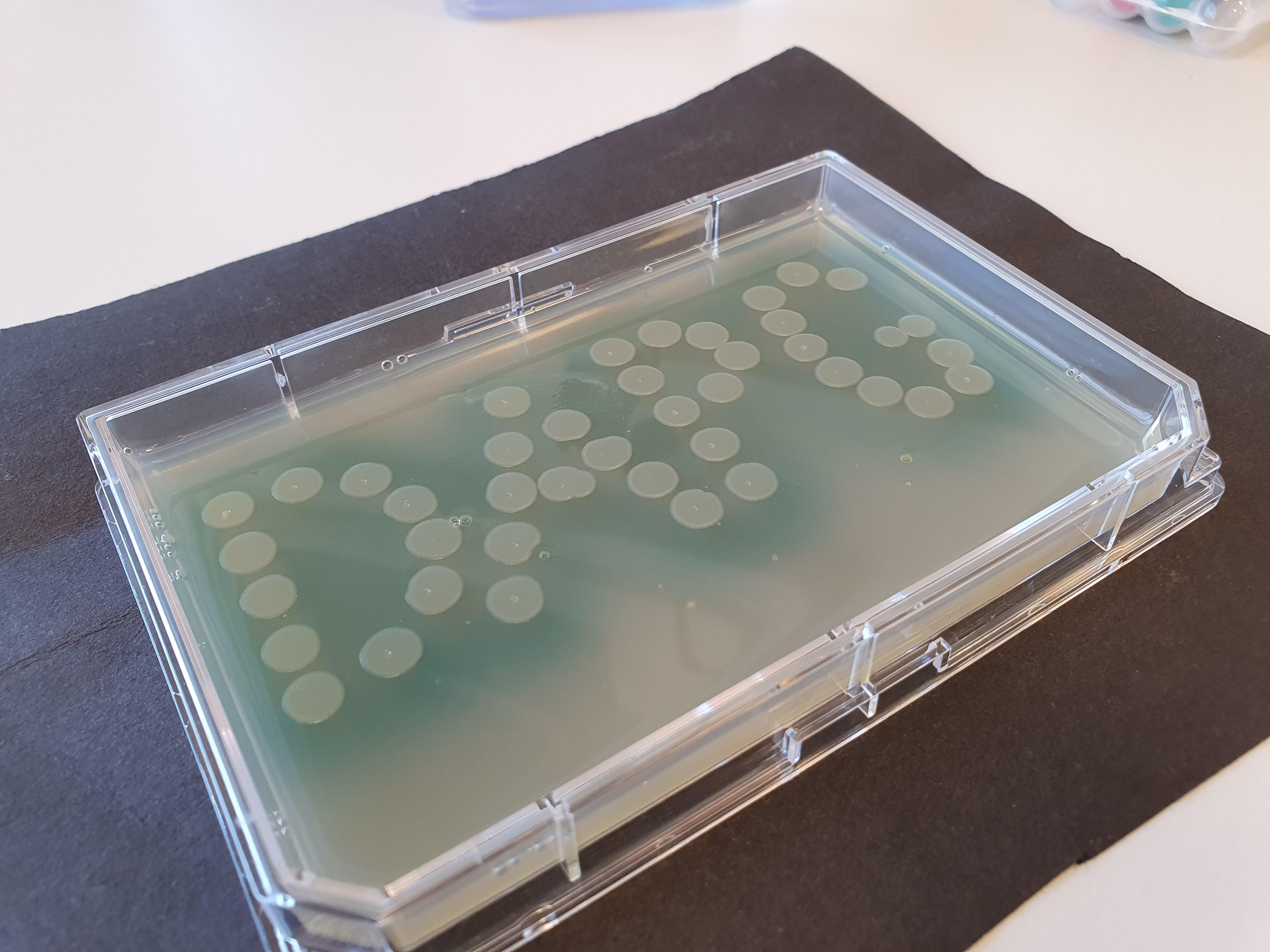
Biosensor that detects low amounts of the bacterial secondary metabolite diacetylphloroglucinol (DAPG). The biosensor consists of a circular piece of DNA (a plasmid) with a repressor module that, upon the presence of DAPG, allows the transcription of a reporter gene giving rise to a detectable response. In the absence of DAPG the repressor blocks transcription thus rendering the biosensor unresponsive. The biosensor has been used to screen the abundance of DAPG producing Pseudomonas species in soil from Danish grassland.

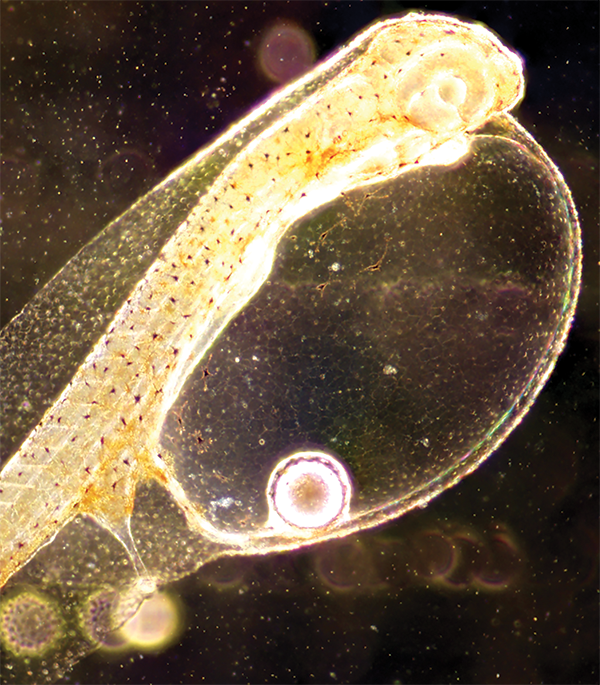
As in CeMiSt, PhD student Karen Dittmann asked what impact a secondary metabolite producing marine bacterium would have on the composition of these microbiomes. The bacterium, Phaeobacter inhibens, produces a potent antimicrobial secondary metabolite, tropodithietic acid (TDA). Whilst it is known that TDA in its pure form has antibiotic activity, it is not currently known how the natural producer affects host-associated microbial communities. Using DNA sequencing, Karen found that the composition of the microbiomes were primarily affected in algae and copepods, namely, the bottom layers of the food chain. Here, the presence of TDA-producing P. inhibens reduced the abundance of a group of bacteria called Rhodobacterales. This group is closely related to the TDA-producer and the results indicate that this secondary metabolite may serve as an interacting molecule amongst closely related species.